The new Illumina patterned flow cell technology uses chemistry that is prone to “index hopping”, leading to reads being assigned to the wrong sample in multiplexed sequencing runs. This problem has already caused countless research groups to re-assess their experimental results or refrain from using the new sequencers. However, with careful experimental preparation it may be possible to ameliorate this situation so as to become negligible for most applications
June 7, 2017
Steven Wingett
HiSeq, Illumina, X Ten, All Applications
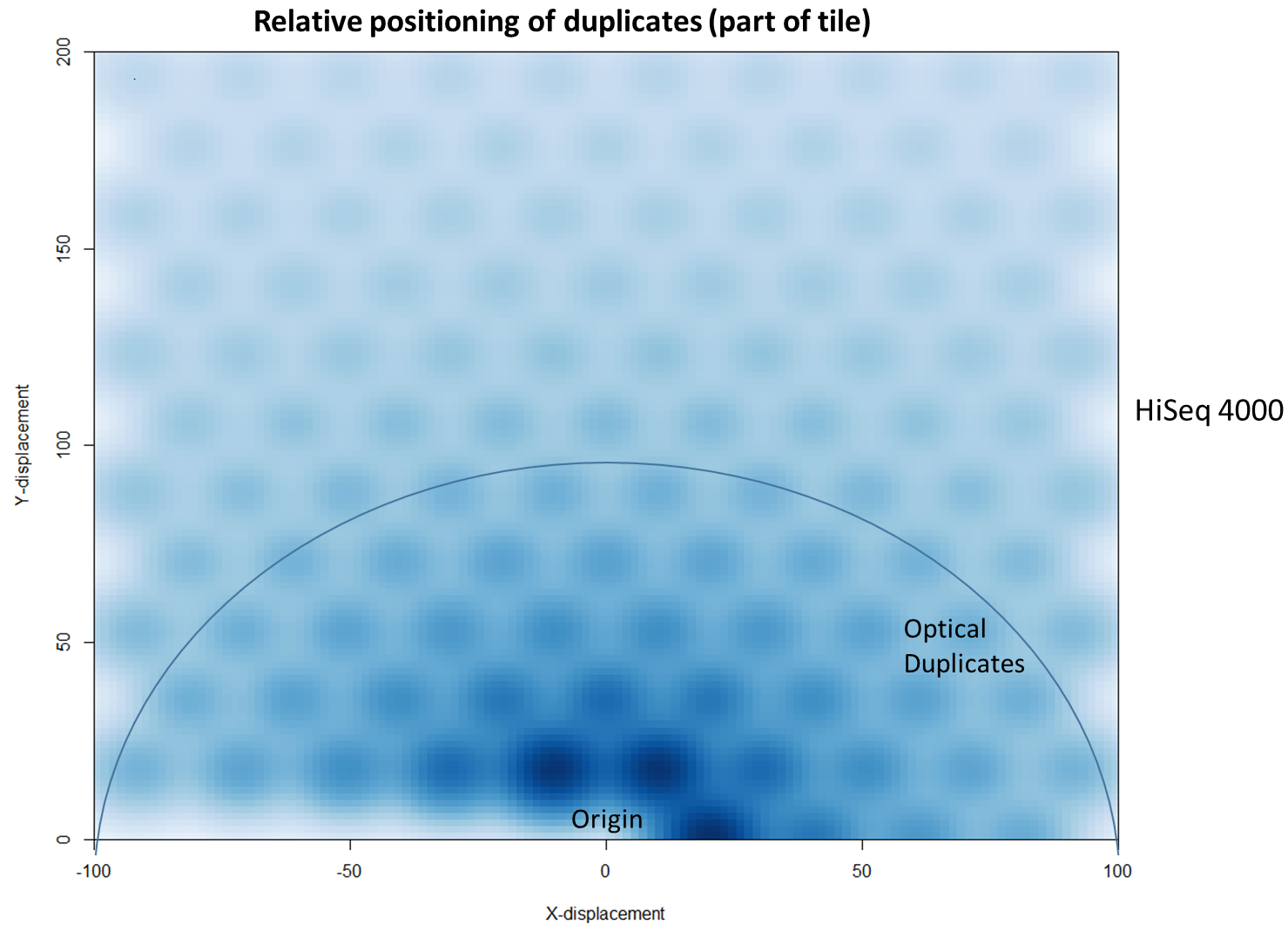
The latest Illumina sequencers – such as the HiSeq X, HiSeq 3000 and HiSeq 4000 – use patterned flow cells to enable the discrimination between much more densely packed DNA clusters. While such technology substantially increases the number of reads generated per sequence run, this innovation may lead to an increased number of duplicates, thereby negating the improved yield and making subsequent data analysis potentially more difficult. Further investigation shows that these putative sequencing duplicates are generally in close two-dimensional proximity on a flow cell, which may provide an opportunity to develop bioinformatics solutions to identify and discard such artefacts.
March 2, 2017
Steven Wingett
HiSeq, All Applications, Bowtie2, HiCUP, Picard
With the introduction of the NextSeq system Illumina changed the way their image data was acquired so that instead of capturing 4 images per cycle they needed only 2. This speeds up image acquisition significantly but also introduces a problem where high quality calls for G bases can be made where there is actually no signal on the flowcell.
May 4, 2016
Simon Andrews
NextSeq, All Applications, Cutadapt, FastQC
With the increasing capacity of a single flowcell lane it can be tempting to mix samples of different types within the same lane to make the most of your sequencing, but cross contamination between libraries in a flowcell can lead to the generation of artefacts which can mess up your analysis.
April 15, 2016
Simon Andrews
Illumina, All Applications, SeqMonk

Paired-end libraries generated by Post Bisulfite Adapter Tagging (PBAT) often suffer from poorer mapping efficiencies when compared to standard whole genome shotgun Bisulfite-Seq libraries. In addition to the usual suspects that have a detrimental impact on mapping efficiency we found that a substantial proportion of paired-end PBAT libraries appears to consist of chimeric reads that map to different places in the genome, not unlike Hi-C type experiments. Chimeric reads also affect single-cell libraries (scBS-seq) as they are constructed using a PBAT approach.
March 18, 2016
Felix Krueger
Illumina, Methylation, PBAT, Bismark, Cutadapt, SeqMonk, Trim Galore!
In some experimental designs a large proportion of the sequences in a library can have identical sequence at their 5′ end. These types of library can cause problems for the data collection and base calling on illumina sequencers, leading to the generation of poor quality data.
March 15, 2016
Simon Andrews
Illumina, All Applications, FastQC
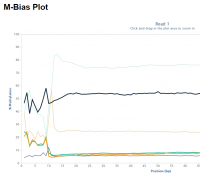
Random priming in PBAT libraries introduces drastic biases in the base composition and methylation levels especially at the 5′ end of all reads. As a result, affected bases should be removed from the libraries before the alignment step.
March 11, 2016
Felix Krueger
Illumina, Methylation, PBAT, BamQC, Bismark, FastQC, Trim Galore!

Library construction of standard directional BS-Seq samples often consist of several steps including sonication, end-repair, A-tailing and adapter ligation. Since the end-repair step typically uses unmethylated cytosines for the fill-in reaction the filled-in bases will generally appear unmethylated after bisulfite conversion irrespective of their true genomic methylation state.
February 12, 2016
Felix Krueger
Illumina, BS-Seq, Methylation, Bismark, Data Processing

The construction of sequencing libraries on many platforms requires the addition of specific adapter sequences to the ends of the fragments to be sequenced. Although there are steps in place to ensure that only valid adapter+insert combinations make it onto the sequencer it is possible to get adapter dimers with no valid insert making it through the sequencing process.
February 1, 2016
Simon Andrews
Illumina, FastQC

Rather than a general loss of quality for a whole sequencing lane or run sometimes partial failures occur. These can affect specific regions and cycles and can have knock on effects for the data generated
January 31, 2016
Simon Andrews
Illumina, FastQC
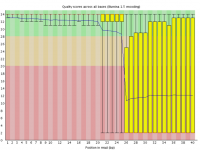
Sometimes a sequencing run can experience a sudden and lasting loss of base call quality across all sequences.
January 20, 2016
Simon Andrews
Illumina, FastQC, QC Software
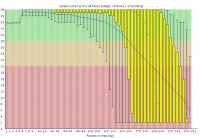
Illumina based sequencing shows a loss of base call quality as the number of sequencing cycles performed increases.
January 20, 2016
Simon Andrews
Illumina, FastQC, QC Software












