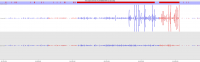
Soft-clipping of reads may add potentially unwanted alignments to repetitive regions
Soft-clipping of sequencing reads allows the masking of portions of the reads that do not align to the genome from end to end, which may be desirable for certain types of analysis (e.g. detection of structural variants). For standard alignment processes soft-clipping may however incorrectly trim reads and lead to the mis-assignments of reads primarily to repetitive regions. This phenomenon appears to vary in severity for different sequencing applications with Bisulfite sequencing being worst off.