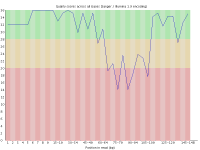
With the introduction of the NextSeq system Illumina changed the way their image data was acquired so that instead of capturing 4 images per cycle they needed only 2. This speeds up image acquisition significantly but also introduces a problem where high quality calls for G bases can be made where there is actually no signal on the flowcell.
May 4, 2016
Simon Andrews
NextSeq, All Applications, Cutadapt, FastQC

Paired-end libraries generated by Post Bisulfite Adapter Tagging (PBAT) often suffer from poorer mapping efficiencies when compared to standard whole genome shotgun Bisulfite-Seq libraries. In addition to the usual suspects that have a detrimental impact on mapping efficiency we found that a substantial proportion of paired-end PBAT libraries appears to consist of chimeric reads that map to different places in the genome, not unlike Hi-C type experiments. Chimeric reads also affect single-cell libraries (scBS-seq) as they are constructed using a PBAT approach.
March 18, 2016
Felix Krueger
Illumina, Methylation, PBAT, Bismark, Cutadapt, SeqMonk, Trim Galore!
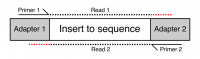
Many sequencing platforms require the addition of specific adapter sequences to the end of the fragments to be sequenced. For an individual fragment, if the length of the sequencing read is longer than the fragment to be sequenced then the read will continue into the adapter sequence on the end. Unless it is removed this adapter sequence will cause problems for downstream mapping, assembly or other analysis.
February 7, 2016
Simon Andrews
Cutadapt, FastQC, Skewer, Trim Galore!




