The new Illumina patterned flow cell technology uses chemistry that is prone to “index hopping”, leading to reads being assigned to the wrong sample in multiplexed sequencing runs. This problem has already caused countless research groups to re-assess their experimental results or refrain from using the new sequencers. However, with careful experimental preparation it may be possible to ameliorate this situation so as to become negligible for most applications
June 7, 2017
Steven Wingett
HiSeq, Illumina, X Ten, All Applications
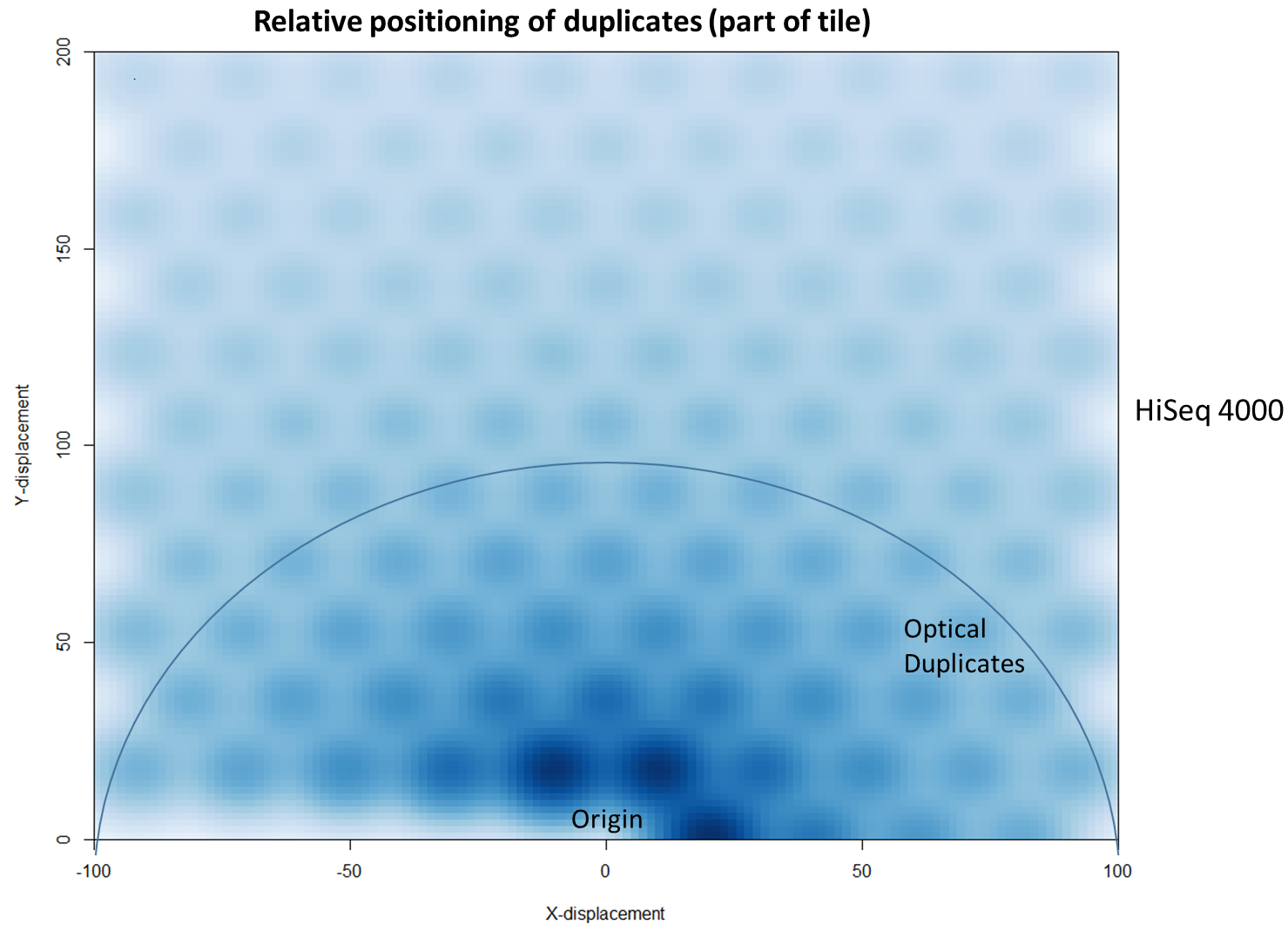
The latest Illumina sequencers – such as the HiSeq X, HiSeq 3000 and HiSeq 4000 – use patterned flow cells to enable the discrimination between much more densely packed DNA clusters. While such technology substantially increases the number of reads generated per sequence run, this innovation may lead to an increased number of duplicates, thereby negating the improved yield and making subsequent data analysis potentially more difficult. Further investigation shows that these putative sequencing duplicates are generally in close two-dimensional proximity on a flow cell, which may provide an opportunity to develop bioinformatics solutions to identify and discard such artefacts.
March 2, 2017
Steven Wingett
HiSeq, All Applications, Bowtie2, HiCUP, Picard


